Topology prediction algorithms
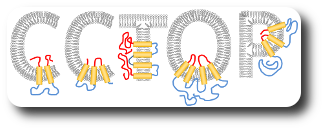
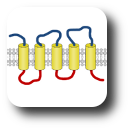 The group leader has been engaged in sequence analysis of transmembrane proteins since the end of the 1990s and achieved remarkable results in the field of topology prediction. In a recent research, we developed an algorithm that incorporates the prediction results of the most accurate state-of-the-art prediction methods as well as experimentally established topology data and other computational biology results. The developed constrained, consensus prediction method (CCTOP) is the most accurate prediction method so far, therefore it was used for creating the human transmembrane proteome (HTP database).
The group leader has been engaged in sequence analysis of transmembrane proteins since the end of the 1990s and achieved remarkable results in the field of topology prediction. In a recent research, we developed an algorithm that incorporates the prediction results of the most accurate state-of-the-art prediction methods as well as experimentally established topology data and other computational biology results. The developed constrained, consensus prediction method (CCTOP) is the most accurate prediction method so far, therefore it was used for creating the human transmembrane proteome (HTP database).
Related publications
-
Dobson L, Szekeres LI, Gedán C, Langó T, Zeke A and Tusnády GE (2023) TmAlphaFold database: membrane localization and evaluation of AlphaFold2 predicted alpha-helical transmembrane protein structures Nucleic Acids Research , .
-
Bakos É, Németh O, Kucsma N, Tőkési N, Stieger B, Rushing E, Tőkés A, Kele P, Tusnády GE and Özvegy-Laczka Cs (2022) Cloning and characterization of a novel functional organic anion transporting polypeptide 3A1 isoform highly expressed in the human brain and testis Frontiers in Pharmacology 13, 958023.
-
Dobson L and Tusnády GE (2021) MemDis: Predicting Disordered Regions in Transmembrane Proteins Int. J. Mol. Sci. 22, 12270.
-
Czimer D, Porok K, Csete D, Gyüre Zs, Lavró V, Fülöp K, Chen Z, Gyergyák H, Tusnády GE, Burgess SM, Mócsai A, Váradi A and Varga M (2021) A New Zebrafish Model for Pseudoxanthoma Elasticum Front Cell Dev Biol 9, 628699.
-
Dobson L, Zeke A and Tusnády GE (2021) PolarProtPred: Predicting apical and basolateral localization of transmembrane proteins using putative short linear motifs and deep learning. Bioinformatics , btab480.
-
Zeke A, Dobson L, Szekeres LI, Langó T and Tusnády GE (2021) PolarProtDb: A Database of Transmembrane and Secreted Proteins showing Apical-Basal Polarity J Mol Biol 433, 166705.
-
Kozák E, Szikora B, Iliás A, Jani PK, Hegyi Z, Matula Zs, Dedinszki D, Tőkési N, Fülöp K, Pomozi V, Várady Gy, Bakos É, Tusnády GE, Kacskovics I and Váradi A (2020) Creation of the first monoclonal antibody recognizing an extracellular epitope of hABCC6 FEBS Lett 595, 789-798.
-
Bakos É, Tusnády GE, Német O, Patik I, Magyar C, Németh K, Kele P and Özvegy- Laczka C (2020) Synergistic transport of a fluorescent coumarin probe marks coumarins as pharmacological modulators of Organic anion-transporting polypeptide, OATP3A1 Biochem Pharmacol. , .
-
Langó T, Pataki ZG, Turiák L, Ács A, Varga JK, Várady, G, Kucsma, N, Drahos, L and Tusnády GE (2020) Partial Proteolysis Improves the Identification of the Extracellular Segments of Transmembrane Proteins by Surface Biotinylation Scientific Reports 10, 8880.
-
Vargja, JK and Tusnady, GE (2019) The TMCrys server for supporting crystallization of transmembrane proteins. Bioinformatics 35, 4203-4.
-
Muller, A, Lango, T, Turiak, L, Acs, A, Varady, G, Kucsma, N, Drahos, L and Tusnady, GE (2019) Covalently modified carboxyl side chains on cell surface leads to a novel method toward topology analysis of transmembrane proteins Scientific Reports 9, 15729.
-
Varga J and Tusnady, GE (2018) TMCrys: predict propensity of success for transmembrane protein crystallization. Bioinformatics , .
-
Lango T, Rona G, Hunyadi-Gulyas E, Turiak L, Varga J, Dobson L, Varady Gy, Drahos L, Vertessy BG, Medzihradszky FK, Szakacs G and Tusnady GE (2017) Identification of extracellular segments by mass spectrometry improves topology prediction of transmembrane proteins Scientific Reports 7, 42610.
-
Varga J, Dobson L, Remenyi I and Tusnady GE (2017) TSTMP: target selection for structural genomics of human transmembrane proteins. Nucleic Acids Res 45, D325-330.
-
Varga J, Dobson L and Tusnady GE (2016) TOPDOM: database of conservatively located domains and motifs in proteins. Bioinformatics 32, 2725-2726.
-
Dobson L, Remenyi I and Tusnady GE (2015) The human transmembrane proteome. Biol Direct 10, 31.
-
Dobson L, Remenyi I and Tusnady GE (2015) CCTOP: a Consensus Constrained TOPology prediction web server. Nucleic Acids Res 43, W408-12.
-
Kiss K, Kucsma N, Brozik A, Tusnady GE, Bergam P, van Niel G and Szakacs G (2015) Role of the N-terminal transmembrane domain in the endo-lysosomal targeting and Biochem J 467, 127-39.
-
Dobson L, Lango T, Remenyi I and Tusnady GE (2015) Expediting topology data gathering for the TOPDB database. Nucleic Acids Res 43, D283-9.
-
Tusnady GE and Simon I (2010) Topology prediction of helical transmembrane proteins: how far have we reached? Curr Protein Pept Sci 11, 550-61.
-
Tusnady GE, Kalmar L, Hegyi H, Tompa P and Simon I (2008) TOPDOM: database of domains and motifs with conservative location in Bioinformatics 24, 1469-70.
-
Tusnady GE, Kalmar L and Simon I (2008) TOPDB: topology data bank of transmembrane proteins. Nucleic Acids Res 36, D234-9.


 The group leader has been engaged in sequence analysis of transmembrane proteins since the end of the 1990s and achieved remarkable results in the field of topology prediction. In a recent research, we developed an algorithm that incorporates the prediction results of the most accurate state-of-the-art prediction methods as well as experimentally established topology data and other computational biology results. The developed constrained, consensus prediction method (CCTOP) is the most accurate prediction method so far, therefore it was used for creating the human transmembrane proteome (HTP database).
The group leader has been engaged in sequence analysis of transmembrane proteins since the end of the 1990s and achieved remarkable results in the field of topology prediction. In a recent research, we developed an algorithm that incorporates the prediction results of the most accurate state-of-the-art prediction methods as well as experimentally established topology data and other computational biology results. The developed constrained, consensus prediction method (CCTOP) is the most accurate prediction method so far, therefore it was used for creating the human transmembrane proteome (HTP database).

